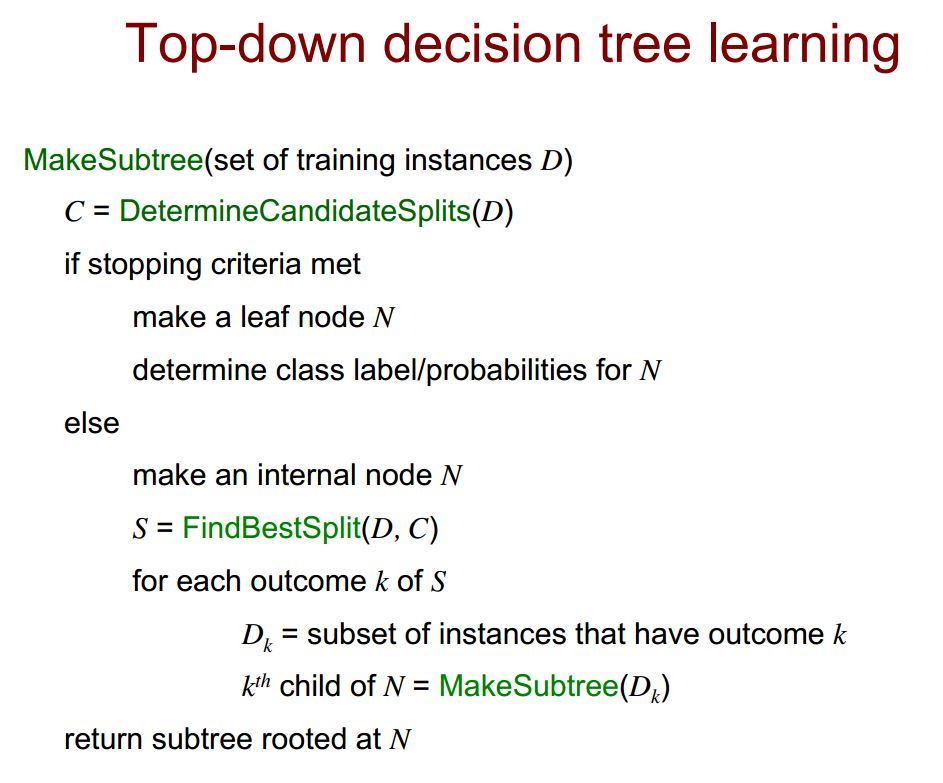
package rdd.ml.classification.tree
import org.apache.spark.mllib.tree.DecisionTree
import org.apache.spark.mllib.tree.model.DecisionTreeModel
import org.apache.spark.mllib.util.MLUtils
import rdd.ml.DFUtils
/**
* 用于分类的决策树模型
* Created by Du Fei on 2018/7/31.
*/
object DecisionTreeForClassification {
def main(args: Array[String]): Unit = {
//这里封装了一个初始化Spark环境的包,可以按照正常的来
val sc = DFUtils.getSparkContext()
// Load and parse the data file. 使用自带的包载入LibSVM格式的数据
val data = MLUtils.loadLibSVMFile(sc, "file:/d:/data/sample_libsvm_data.txt")
// Split the data into training and test sets (30% held out for testing) 将数据集划分成训练集和测试集
val splits = data.randomSplit(Array(0.7, 0.3))
val (trainingData, testData) = (splits(0), splits(1))
// Train a DecisionTree model. 决策树参数
// Empty categoricalFeaturesInfo indicates all features are continuous.
val numClasses = 2 //类标签数量,这是个二分类问题
val categoricalFeaturesInfo = Map[Int, Int]()
val impurity = "gini" //使用gini不纯度计算特征信息量
val maxDepth = 5 //树的最大深度是5
val maxBins = 32 //特征划分结果最多只有5个
// 训练模型
val model = DecisionTree.trainClassifier(trainingData, numClasses, categoricalFeaturesInfo,
impurity, maxDepth, maxBins)
// Evaluate model on test instances and compute test error
val labelAndPreds = testData.map { point =>
val prediction = model.predict(point.features)
(point.label, prediction)
}
val testErr = labelAndPreds.filter(r => r._1 != r._2).count().toDouble / testData.count()
println(s"Test Error = $testErr")
println(s"Learned classification tree model:\n ${model.toDebugString}")
// Save and load model
model.save(sc, "target/tmp/myDecisionTreeClassificationModel")
val sameModel = DecisionTreeModel.load(sc, "target/tmp/myDecisionTreeClassificationModel")
}
}
@Since("1.1.0")
def trainClassifier(
input: RDD[LabeledPoint],
numClasses: Int,
categoricalFeaturesInfo: Map[Int, Int],
impurity: String,
maxDepth: Int,
maxBins: Int): DecisionTreeModel = {
// 设置信息量计算方式
val impurityType = Impurities.fromString(impurity)
// 封装策略
val strategy = new Strategy(algo, impurity, maxDepth, numClasses, maxBins,
quantileCalculationStrategy, categoricalFeaturesInfo)
// 调用随机森林算法训练
val rf = new RandomForest(strategy, numTrees = 1, featureSubsetStrategy = "all", seed = seed)
val rfModel = rf.run(input)
rfModel.trees(0) //将第一棵树返回即可得到决策树训练结果
}
/**
* Train a random forest.
*
* @param input Training data: RDD of `LabeledPoint`
* @return an unweighted set of trees
*/
def run(
input: RDD[LabeledPoint],
strategy: OldStrategy,
numTrees: Int,
featureSubsetStrategy: String,
seed: Long,
instr: Option[Instrumentation[_]],
parentUID: Option[String] = None): Array[DecisionTreeModel] = {
// timer是用来记录运行时间的,可以忽略
val timer = new TimeTracker()
timer.start("total")
timer.start("init")
// retag是给数据重新打标签,为了和Java相兼容而执行的,如果不加这个,在和java通信的时候可能会有问题
val retaggedInput = input.retag(classOf[LabeledPoint])
// 步骤一:构造决策树的元数据,这里面大多数和strategy都重复了,感觉实际上没必要搞这么复杂
val metadata =
DecisionTreeMetadata.buildMetadata(retaggedInput, strategy, numTrees, featureSubsetStrategy)
// 监控
instr match {
case Some(instrumentation) =>
instrumentation.logNumFeatures(metadata.numFeatures)
instrumentation.logNumClasses(metadata.numClasses)
case None =>
logInfo("numFeatures: " + metadata.numFeatures)
logInfo("numClasses: " + metadata.numClasses)
}
// Find the splits and the corresponding bins (interval between the splits) using a sample
// of the input data.
timer.start("findSplits")
// 步骤二:对数据集的特征进行离散化(装箱),主要是针对连续属性来操作,还有无序分类属性以及有序分类属性三种)
// 参考:https://www.datalearner.com/blog/1051533040913424
val splits = findSplits(retaggedInput, metadata, seed)
// 这几句都是日志类的,不用看
timer.stop("findSplits")
logDebug("numBins: feature: number of bins")
logDebug(Range(0, metadata.numFeatures).map { featureIndex =>
s"\t$featureIndex\t${metadata.numBins(featureIndex)}"
}.mkString("\n"))
// Bin feature values (TreePoint representation).
// Cache input RDD for speedup during multiple passes.
val treeInput = TreePoint.convertToTreeRDD(retaggedInput, splits, metadata)
val withReplacement = numTrees > 1
val baggedInput = BaggedPoint
.convertToBaggedRDD(treeInput, strategy.subsamplingRate, numTrees, withReplacement, seed)
.persist(StorageLevel.MEMORY_AND_DISK)
// depth of the decision tree 这里有一个限制,就是树的深度不能超过30,可能是因为性能
val maxDepth = strategy.maxDepth
require(maxDepth <= 30,
s"DecisionTree currently only supports maxDepth <= 30, but was given maxDepth = $maxDepth.")
// Max memory usage for aggregates 内存使用限制
// TODO: Calculate memory usage more precisely.
val maxMemoryUsage: Long = strategy.maxMemoryInMB * 1024L * 1024L
logDebug("max memory usage for aggregates = " + maxMemoryUsage + " bytes.")
/*
* The main idea here is to perform group-wise training of the decision tree nodes thus
* reducing the passes over the data from (# nodes) to (# nodes / maxNumberOfNodesPerGroup).
* Each data sample is handled by a particular node (or it reaches a leaf and is not used
* in lower levels).
*/
// Create an RDD of node Id cache.
// At first, all the rows belong to the root nodes (node Id == 1).
val nodeIdCache = if (strategy.useNodeIdCache) {
Some(NodeIdCache.init(
data = baggedInput,
numTrees = numTrees,
checkpointInterval = strategy.checkpointInterval,
initVal = 1))
} else {
None
}
/*
Stack of nodes to train: (treeIndex, node)
The reason this is a stack is that we train many trees at once, but we want to focus on
completing trees, rather than training all simultaneously. If we are splitting nodes from
1 tree, then the new nodes to split will be put at the top of this stack, so we will continue
training the same tree in the next iteration. This focus allows us to send fewer trees to
workers on each iteration; see topNodesForGroup below.
*/
// 用一个可变的数组堆栈来存储需要训练的节点,里面放的是树的索引和节点LearningNode,由于这里说的是决策树,所以树只有一个
val nodeStack = new mutable.ArrayStack[(Int, LearningNode)]
val rng = new Random()
rng.setSeed(seed)
// Allocate and queue root nodes.
// 把节点放到前面的数组堆栈中,这里只有一个节点
val topNodes = Array.fill[LearningNode](numTrees)(LearningNode.emptyNode(nodeIndex = 1))
Range(0, numTrees).foreach(treeIndex => nodeStack.push((treeIndex, topNodes(treeIndex))))
timer.stop("init")
// 步骤三: 循环训练所有的节点,由于是决策树,这里只会循环一次
while (nodeStack.nonEmpty) {
// Collect some nodes to split, and choose features for each node (if subsampling).
// Each group of nodes may come from one or multiple trees, and at multiple levels.
// 步骤3.1:选择某些节点进行划分,决策树没得节点可选。
val (nodesForGroup, treeToNodeToIndexInfo) =
RandomForest.selectNodesToSplit(nodeStack, maxMemoryUsage, metadata, rng)
// Sanity check (should never occur):
assert(nodesForGroup.nonEmpty,
s"RandomForest selected empty nodesForGroup. Error for unknown reason.")
// Only send trees to worker if they contain nodes being split this iteration.
val topNodesForGroup: Map[Int, LearningNode] =
nodesForGroup.keys.map(treeIdx => treeIdx -> topNodes(treeIdx)).toMap
// Choose node splits, and enqueue new nodes as needed.
timer.start("findBestSplits")
// 为当前的树寻找最佳的划分
RandomForest.findBestSplits(baggedInput, metadata, topNodesForGroup, nodesForGroup,
treeToNodeToIndexInfo, splits, nodeStack, timer, nodeIdCache)
timer.stop("findBestSplits")
}
baggedInput.unpersist()
timer.stop("total")
logInfo("Internal timing for DecisionTree:")
logInfo(s"$timer")
// Delete any remaining checkpoints used for node Id cache.
if (nodeIdCache.nonEmpty) {
try {
nodeIdCache.get.deleteAllCheckpoints()
} catch {
case e: IOException =>
logWarning(s"delete all checkpoints failed. Error reason: ${e.getMessage}")
}
}
val numFeatures = metadata.numFeatures
parentUID match {
case Some(uid) =>
if (strategy.algo == OldAlgo.Classification) {
topNodes.map { rootNode =>
new DecisionTreeClassificationModel(uid, rootNode.toNode, numFeatures,
strategy.getNumClasses)
}
} else {
topNodes.map { rootNode =>
new DecisionTreeRegressionModel(uid, rootNode.toNode, numFeatures)
}
}
case None =>
if (strategy.algo == OldAlgo.Classification) {
topNodes.map { rootNode =>
new DecisionTreeClassificationModel(rootNode.toNode, numFeatures,
strategy.getNumClasses)
}
} else {
topNodes.map(rootNode => new DecisionTreeRegressionModel(rootNode.toNode, numFeatures))
}
}
}
/**
* Given a group of nodes, this finds the best split for each node.
*
* @param input Training data: RDD of [[TreePoint]]
* @param metadata Learning and dataset metadata
* @param topNodesForGroup For each tree in group, tree index -> root node.
* Used for matching instances with nodes.
* @param nodesForGroup Mapping: treeIndex --> nodes to be split in tree
* @param treeToNodeToIndexInfo Mapping: treeIndex --> nodeIndex --> nodeIndexInfo,
* where nodeIndexInfo stores the index in the group and the
* feature subsets (if using feature subsets).
* @param splits possible splits for all features, indexed (numFeatures)(numSplits)
* @param nodeStack Queue of nodes to split, with values (treeIndex, node).
* Updated with new non-leaf nodes which are created.
* @param nodeIdCache Node Id cache containing an RDD of Array[Int] where
* each value in the array is the data point's node Id
* for a corresponding tree. This is used to prevent the need
* to pass the entire tree to the executors during
* the node stat aggregation phase.
*/
private[tree] def findBestSplits(
input: RDD[BaggedPoint[TreePoint]],
metadata: DecisionTreeMetadata,
topNodesForGroup: Map[Int, LearningNode],
nodesForGroup: Map[Int, Array[LearningNode]],
treeToNodeToIndexInfo: Map[Int, Map[Int, NodeIndexInfo]],
splits: Array[Array[Split]],
nodeStack: mutable.ArrayStack[(Int, LearningNode)],
timer: TimeTracker = new TimeTracker,
nodeIdCache: Option[NodeIdCache] = None): Unit = {
/*
* The high-level descriptions of the best split optimizations are noted here.
*
* *Group-wise training*
* We perform bin calculations for groups of nodes to reduce the number of
* passes over the data. Each iteration requires more computation and storage,
* but saves several iterations over the data.
*
* *Bin-wise computation*
* We use a bin-wise best split computation strategy instead of a straightforward best split
* computation strategy. Instead of analyzing each sample for contribution to the left/right
* child node impurity of every split, we first categorize each feature of a sample into a
* bin. We exploit this structure to calculate aggregates for bins and then use these aggregates
* to calculate information gain for each split.
*
* *Aggregation over partitions*
* Instead of performing a flatMap/reduceByKey operation, we exploit the fact that we know
* the number of splits in advance. Thus, we store the aggregates (at the appropriate
* indices) in a single array for all bins and rely upon the RDD aggregate method to
* drastically reduce the communication overhead.
*/
// numNodes: Number of nodes in this group
val numNodes = nodesForGroup.values.map(_.length).sum
logDebug("numNodes = " + numNodes)
logDebug("numFeatures = " + metadata.numFeatures)
logDebug("numClasses = " + metadata.numClasses)
logDebug("isMulticlass = " + metadata.isMulticlass)
logDebug("isMulticlassWithCategoricalFeatures = " +
metadata.isMulticlassWithCategoricalFeatures)
logDebug("using nodeIdCache = " + nodeIdCache.nonEmpty.toString)
/**
* Performs a sequential aggregation over a partition for a particular tree and node.
*
* For each feature, the aggregate sufficient statistics are updated for the relevant
* bins.
*
* @param treeIndex Index of the tree that we want to perform aggregation for.
* @param nodeInfo The node info for the tree node.
* @param agg Array storing aggregate calculation, with a set of sufficient statistics
* for each (node, feature, bin).
* @param baggedPoint Data point being aggregated.
*/
def nodeBinSeqOp(
treeIndex: Int,
nodeInfo: NodeIndexInfo,
agg: Array[DTStatsAggregator],
baggedPoint: BaggedPoint[TreePoint]): Unit = {
if (nodeInfo != null) {
val aggNodeIndex = nodeInfo.nodeIndexInGroup
val featuresForNode = nodeInfo.featureSubset
val instanceWeight = baggedPoint.subsampleWeights(treeIndex)
if (metadata.unorderedFeatures.isEmpty) {
orderedBinSeqOp(agg(aggNodeIndex), baggedPoint.datum, instanceWeight, featuresForNode)
} else {
mixedBinSeqOp(agg(aggNodeIndex), baggedPoint.datum, splits,
metadata.unorderedFeatures, instanceWeight, featuresForNode)
}
agg(aggNodeIndex).updateParent(baggedPoint.datum.label, instanceWeight)
}
}
/**
* Performs a sequential aggregation over a partition.
*
* Each data point contributes to one node. For each feature,
* the aggregate sufficient statistics are updated for the relevant bins.
*
* @param agg Array storing aggregate calculation, with a set of sufficient statistics for
* each (node, feature, bin).
* @param baggedPoint Data point being aggregated.
* @return agg
*/
def binSeqOp(
agg: Array[DTStatsAggregator],
baggedPoint: BaggedPoint[TreePoint]): Array[DTStatsAggregator] = {
treeToNodeToIndexInfo.foreach { case (treeIndex, nodeIndexToInfo) =>
val nodeIndex =
topNodesForGroup(treeIndex).predictImpl(baggedPoint.datum.binnedFeatures, splits)
nodeBinSeqOp(treeIndex, nodeIndexToInfo.getOrElse(nodeIndex, null), agg, baggedPoint)
}
agg
}
/**
* Do the same thing as binSeqOp, but with nodeIdCache.
*/
def binSeqOpWithNodeIdCache(
agg: Array[DTStatsAggregator],
dataPoint: (BaggedPoint[TreePoint], Array[Int])): Array[DTStatsAggregator] = {
treeToNodeToIndexInfo.foreach { case (treeIndex, nodeIndexToInfo) =>
val baggedPoint = dataPoint._1
val nodeIdCache = dataPoint._2
val nodeIndex = nodeIdCache(treeIndex)
nodeBinSeqOp(treeIndex, nodeIndexToInfo.getOrElse(nodeIndex, null), agg, baggedPoint)
}
agg
}
/**
* Get node index in group --> features indices map,
* which is a short cut to find feature indices for a node given node index in group.
*/
def getNodeToFeatures(
treeToNodeToIndexInfo: Map[Int, Map[Int, NodeIndexInfo]]): Option[Map[Int, Array[Int]]] = {
if (!metadata.subsamplingFeatures) {
None
} else {
val mutableNodeToFeatures = new mutable.HashMap[Int, Array[Int]]()
treeToNodeToIndexInfo.values.foreach { nodeIdToNodeInfo =>
nodeIdToNodeInfo.values.foreach { nodeIndexInfo =>
assert(nodeIndexInfo.featureSubset.isDefined)
mutableNodeToFeatures(nodeIndexInfo.nodeIndexInGroup) = nodeIndexInfo.featureSubset.get
}
}
Some(mutableNodeToFeatures.toMap)
}
}
// array of nodes to train indexed by node index in group
val nodes = new Array[LearningNode](numNodes)
nodesForGroup.foreach { case (treeIndex, nodesForTree) =>
nodesForTree.foreach { node =>
nodes(treeToNodeToIndexInfo(treeIndex)(node.id).nodeIndexInGroup) = node
}
}
// Calculate best splits for all nodes in the group
timer.start("chooseSplits")
// In each partition, iterate all instances and compute aggregate stats for each node,
// yield a (nodeIndex, nodeAggregateStats) pair for each node.
// After a `reduceByKey` operation,
// stats of a node will be shuffled to a particular partition and be combined together,
// then best splits for nodes are found there.
// Finally, only best Splits for nodes are collected to driver to construct decision tree.
val nodeToFeatures = getNodeToFeatures(treeToNodeToIndexInfo)
val nodeToFeaturesBc = input.sparkContext.broadcast(nodeToFeatures)
val partitionAggregates: RDD[(Int, DTStatsAggregator)] = if (nodeIdCache.nonEmpty) {
input.zip(nodeIdCache.get.nodeIdsForInstances).mapPartitions { points =>
// Construct a nodeStatsAggregators array to hold node aggregate stats,
// each node will have a nodeStatsAggregator
val nodeStatsAggregators = Array.tabulate(numNodes) { nodeIndex =>
val featuresForNode = nodeToFeaturesBc.value.map { nodeToFeatures =>
nodeToFeatures(nodeIndex)
}
new DTStatsAggregator(metadata, featuresForNode)
}
// iterator all instances in current partition and update aggregate stats
points.foreach(binSeqOpWithNodeIdCache(nodeStatsAggregators, _))
// transform nodeStatsAggregators array to (nodeIndex, nodeAggregateStats) pairs,
// which can be combined with other partition using `reduceByKey`
nodeStatsAggregators.view.zipWithIndex.map(_.swap).iterator
}
} else {
input.mapPartitions { points =>
// Construct a nodeStatsAggregators array to hold node aggregate stats,
// each node will have a nodeStatsAggregator
val nodeStatsAggregators = Array.tabulate(numNodes) { nodeIndex =>
val featuresForNode = nodeToFeaturesBc.value.flatMap { nodeToFeatures =>
Some(nodeToFeatures(nodeIndex))
}
new DTStatsAggregator(metadata, featuresForNode)
}
// iterator all instances in current partition and update aggregate stats
points.foreach(binSeqOp(nodeStatsAggregators, _))
// transform nodeStatsAggregators array to (nodeIndex, nodeAggregateStats) pairs,
// which can be combined with other partition using `reduceByKey`
nodeStatsAggregators.view.zipWithIndex.map(_.swap).iterator
}
}
val nodeToBestSplits = partitionAggregates.reduceByKey((a, b) => a.merge(b)).map {
case (nodeIndex, aggStats) =>
val featuresForNode = nodeToFeaturesBc.value.flatMap { nodeToFeatures =>
Some(nodeToFeatures(nodeIndex))
}
// find best split for each node
val (split: Split, stats: ImpurityStats) =
binsToBestSplit(aggStats, splits, featuresForNode, nodes(nodeIndex))
(nodeIndex, (split, stats))
}.collectAsMap()
timer.stop("chooseSplits")
val nodeIdUpdaters = if (nodeIdCache.nonEmpty) {
Array.fill[mutable.Map[Int, NodeIndexUpdater]](
metadata.numTrees)(mutable.Map[Int, NodeIndexUpdater]())
} else {
null
}
// Iterate over all nodes in this group.
nodesForGroup.foreach { case (treeIndex, nodesForTree) =>
nodesForTree.foreach { node =>
val nodeIndex = node.id
val nodeInfo = treeToNodeToIndexInfo(treeIndex)(nodeIndex)
val aggNodeIndex = nodeInfo.nodeIndexInGroup
val (split: Split, stats: ImpurityStats) =
nodeToBestSplits(aggNodeIndex)
logDebug("best split = " + split)
// Extract info for this node. Create children if not leaf.
val isLeaf =
(stats.gain <= 0) || (LearningNode.indexToLevel(nodeIndex) == metadata.maxDepth)
node.isLeaf = isLeaf
node.stats = stats
logDebug("Node = " + node)
if (!isLeaf) {
node.split = Some(split)
val childIsLeaf = (LearningNode.indexToLevel(nodeIndex) + 1) == metadata.maxDepth
val leftChildIsLeaf = childIsLeaf || (stats.leftImpurity == 0.0)
val rightChildIsLeaf = childIsLeaf || (stats.rightImpurity == 0.0)
node.leftChild = Some(LearningNode(LearningNode.leftChildIndex(nodeIndex),
leftChildIsLeaf, ImpurityStats.getEmptyImpurityStats(stats.leftImpurityCalculator)))
node.rightChild = Some(LearningNode(LearningNode.rightChildIndex(nodeIndex),
rightChildIsLeaf, ImpurityStats.getEmptyImpurityStats(stats.rightImpurityCalculator)))
if (nodeIdCache.nonEmpty) {
val nodeIndexUpdater = NodeIndexUpdater(
split = split,
nodeIndex = nodeIndex)
nodeIdUpdaters(treeIndex).put(nodeIndex, nodeIndexUpdater)
}
// enqueue left child and right child if they are not leaves
if (!leftChildIsLeaf) {
nodeStack.push((treeIndex, node.leftChild.get))
}
if (!rightChildIsLeaf) {
nodeStack.push((treeIndex, node.rightChild.get))
}
logDebug("leftChildIndex = " + node.leftChild.get.id +
", impurity = " + stats.leftImpurity)
logDebug("rightChildIndex = " + node.rightChild.get.id +
", impurity = " + stats.rightImpurity)
}
}
}
if (nodeIdCache.nonEmpty) {
// Update the cache if needed.
nodeIdCache.get.updateNodeIndices(input, nodeIdUpdaters, splits)
}
}